Verdonschot JAJ. Eur Heart J. 2021 Jan 7;42(2):162-174.
The dilated cardiomyopathy (DCM) phenotype is the consequence of combined genetic and obtained triggers. Still, clinical decision-making in DCM has primarily been based on ejection fraction (EF) and New York Heart Association (NYHA) classification, not contemplating the DCM heterogenicity. Phenomapping can assist to establish homogeneous DCM subgroups, called phenogroups, based on unsupervised clustering of clinical data. Machine learning helps in diagnosing patterns among variables which describe the heterogeneity in a dataset. The methodology has priory aided to develop clinically valid phenogroups in heart failure with preserved and decreased EF. Until now, in patients with DCM, studies applying machine learning to develop phenogroups are lacking. Thus, Verdonschot JAJ, et al., conducted a study (i) to recognise patterns in clinical data and cluster patients depending on phenotypic similarities; (ii) to study their fundamental molecular profiles and possible pathomechanisms by executing RNA-sequencing on cardiac biopsies of patients from each phenogroup; and (iii) to develop a clinical classifier depending on the created patients clusters and assess its applicability in external DCM groups.
795 consecutive DCM patients were enlisted from the Maastricht Cardiomyopathy Registry in 2006 to 2019, presented with a left ventricular (LV) EF <50% at baseline echocardiographic analysis in the absence of any of the following conditions: obstruction >50% of a major coronary artery branch [at coronary angiography (CAG)], pericardial diseases, congenital heart diseases, cor pulmonale, and active myocarditis. All patients has gone through physical examination, blood sampling, 12-lead electrocardiogram (ECG), 24-h ECG Holter monitoring, a complete echocardiographic and Doppler analysis at baseline and in structured, systematic follow-up. Genetic testing was executed by a 23 DCM-correlated gene panel. A CAG was systematically executed in patients ≥35 years, with cardiovascular risk factors and/or without familial history for DCM in order to exclude ischaemic heart disease. All patients underwent indepth phenotyping, including extensive clinical data on aetiology and comorbidities, imaging and endomyocardial biopsies (EMB).
The data processing of the clinical variables was summarized in Figure 1, selecting 28 variables as input for the cluster analysis.
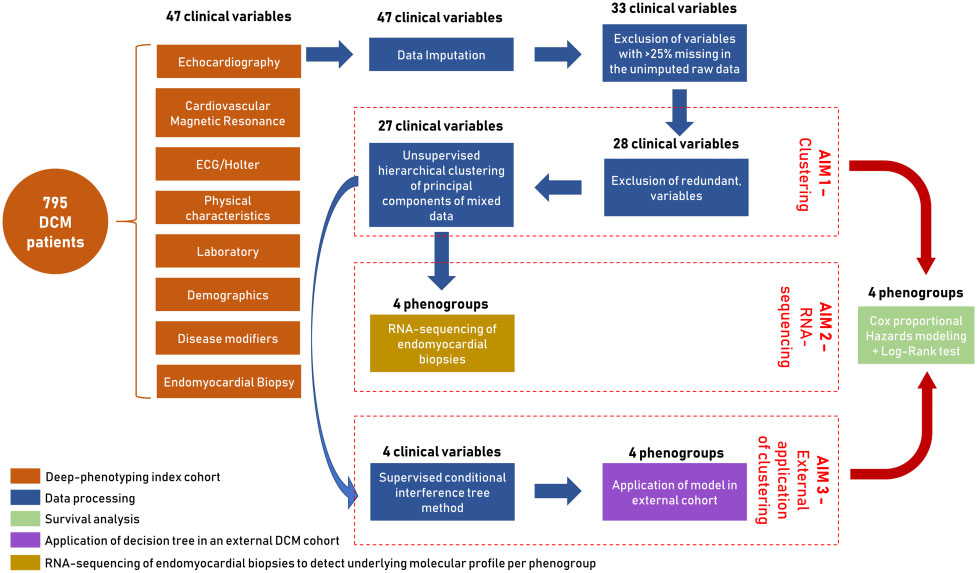
Figure 1: Summary of the aims and study design including data processing steps, survival analysis, and application
4 was the minimal number of clusters which could precisely reflect the phenotypic difference in the index group. 27 of the 28 clinical input variables showed a contribution in describing the patient clusters except the cardiac viral load in EMBs. This resulted in four phenogroups being identified with major variations in their clinical characteristics; [PG1] mild systolic dysfunction, [PG2] auto-immune, [PG3] genetic and arrhythmias, and [PG4] severe systolic dysfunction (Figure 2).
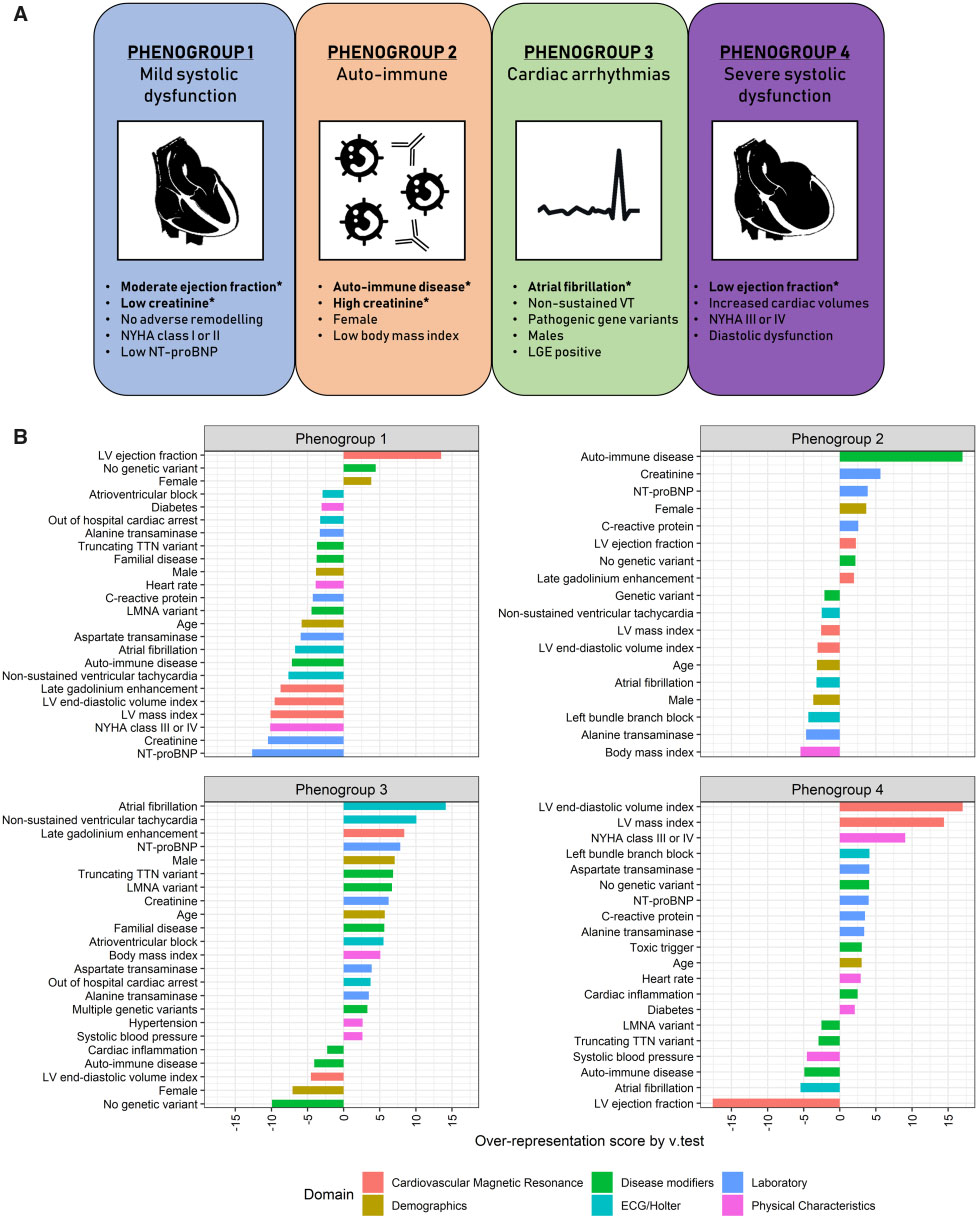
Figure 2: Four mutually exclusive phenogroups as determined by hierarchical clustering of principal component using phenotypical information as input.
The most distinct clinical characteristics are listed per phenogroup. Variables with an asterisk are key parameters to distinguish the phenogroups, as selected by supervised decision tree modelling (A). Characteristic plots of the four proposed phenogroups including their most representative clinical variables. The over- or underrepresentation of a variable within a cluster was analysed by v-test within the hierarchical clustering of principal component function, based on the hypergeometric distribution. A positive value indicates overrepresentation of this variable in the applicable phenogroup, a negative value indicates underrepresentation of the corresponding variable (B).
A genome-wide transcriptome evaluation (RNA-sequencing of EMB) was executed in a sub-cohort of patients with available spare biopsies for RNA-sequencing (n=91, distribution phenogroups 1–4 = 21/9/35/26) to obtain better vision into the cardiac pathophysiological differences among the phenogroups. All phenogroups showed a definite transcriptomic signature isolating them from the other groups. PG4 showed large differences in gene expression, as indicated through the number of differentially expressed genes and strong separation. Subsequent gene set enrichment evaluation with KEGG terms showed unique expression of molecular pathways per phenogroup. Altogether, cardiac metabolism was the potent differentially expressed biological pathway in PG4 [reflecting (i) a reduction in fatty acid pathways and an improvement in pathways included in glycolytic substrate usage; and (ii) an increase in purine and pyrimidine metabolism reflecting DNA replication], PG2 (auto-immune) showed a proinflammatory gene profile (NFKB- and TNF-signalling) and PG3 (arrhythmia) was the most pro-fibrotic (focal adhesion and extracellular matrix remodelling). (Figure 3).
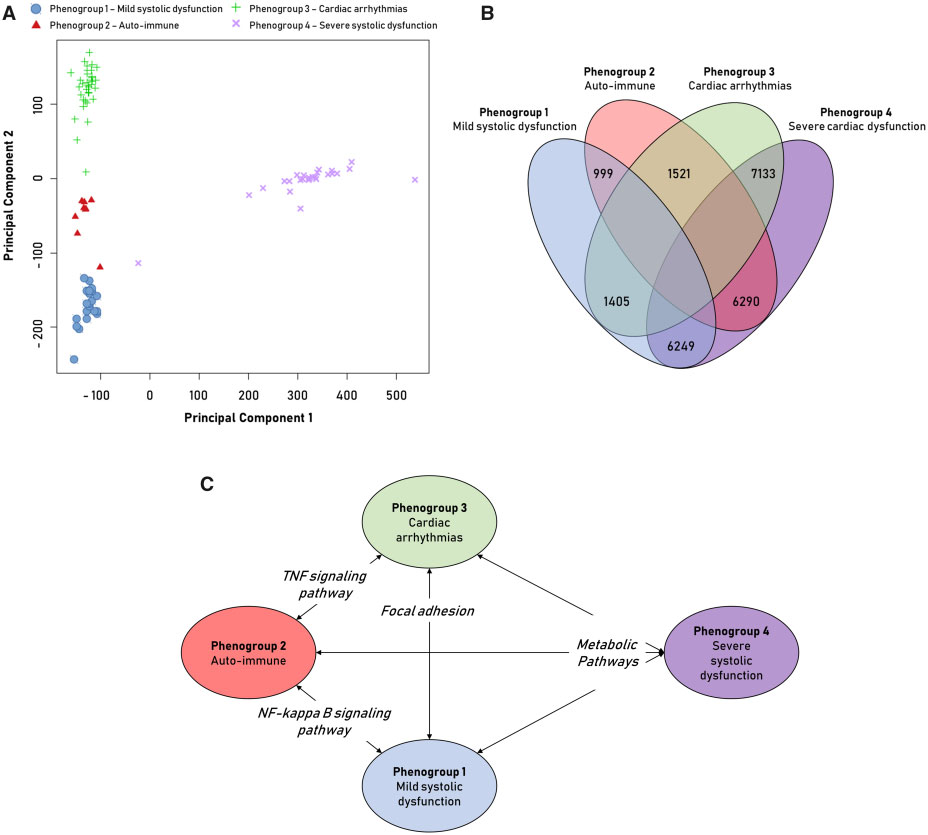
Figure 3: Analysis of RNA-sequencing data of endomyocardial biopsies from dilated cardiomyopathy patients.
Principal component analysis (PCA) of RNA-sequencing data divided on phenogroup (PG). Principal component 1 shows strong division of PG4 and the others (A). Venn diagram of the number of significant differentially expressed genes in the comparison between two corresponding PG (FDR< 0.01þfold change >1.5) (B). Significantly enriched Kyoto Encyclopaedia of Genes andGenomes pathways (P-value< 0.05) in the comparison between the PG (C).
Additionally, event-free survival varied between the four phenogroups, also when rectified for well-known clinical predictors. The presence of an auto-immune disease, LVEF, AF, and creatinine were preferred as the most significant variables characteristic for these phenogroups with a combined precision of 71%.
Thus, it was concluded that the present study recognised four different DCM phenogroups correlated with substantial differences in clinical presentation, fundamental molecular profiles and effect, covering the way for a more personalized approach to therapy.
TNF: Tumor necrosis factor; AF: Atrial fibrillation; LVEF: Left ventricular ejection fraction

